Usage#
Pymech is a simple interface to read / to write Nek5000 and SIMSON specific data files to / from the Python world. With this capability you could:
make publication-quality figures
post-process
manipulate meshes
generate initial conditions
interpolate or extrapolate solution fields, and
possibly many more potential use-cases – the limit is your imagination! Here we look at some simple operations you can do. Start by installing pymech and:
!git clone --recursive https://github.com/eX-Mech/pymech.git
%cd pymech/tests/data/nek
Cloning into 'pymech'...
remote: Enumerating objects: 3041, done.
remote: Counting objects: 100% (692/692), done.
remote: Compressing objects: 100% (218/218), done.
remote: Total 3041 (delta 557), reused 482 (delta 474), pack-reused 2349
Receiving objects: 100% (3041/3041), 12.73 MiB | 33.69 MiB/s, done.
Resolving deltas: 100% (2067/2067), done.
Submodule 'data' (https://github.com/eX-Mech/pymech-test-data.git) registered for path 'tests/data'
Cloning into '/tmp/tmpyvoe0k5g/pymech/tests/data'...
remote: Enumerating objects: 233, done.
remote: Counting objects: 100% (233/233), done.
remote: Compressing objects: 100% (151/151), done.
remote: Total 233 (delta 81), reused 232 (delta 80), pack-reused 0
Receiving objects: 100% (233/233), 25.70 MiB | 34.40 MiB/s, done.
Resolving deltas: 100% (81/81), done.
Submodule path 'tests/data': checked out '9908c266c7d28ee667a6337ee3cfe73ba577fe3e'
/tmp/tmpyvoe0k5g/pymech/tests/data/nek
/home/docs/checkouts/readthedocs.org/user_builds/pymech/envs/latest/lib/python3.9/site-packages/IPython/core/magics/osm.py:417: UserWarning: using dhist requires you to install the `pickleshare` library.
self.shell.db['dhist'] = compress_dhist(dhist)[-100:]
pymech.neksuite#
from pymech.neksuite import readnek
field = readnek('channel3D_0.f00001')
Simply typing the read field would give you some basic information:
field
<pymech.core.HexaData>
Dimensions: 3
Precision: 4 bytes
Mesh limits:
* x: (0.0, 6.2831854820251465)
* y: (-1.0, 1.0)
* z: (0.0, 3.1415927410125732)
Time:
* time: 0.2
* istep: 10
Elements:
* nel: 512
* elem: [<elem centered at [ 0.39269908 -0.98 0.19634954]>
...
<elem centered at [5.89048618 0.98 2.94524309]>]
pymech.core.HexaData#
The pymech.core.HexaData class is the in-memory data structure widely used in Pymech. It stores a description of the field and other metadata. Let’s look at the available attributes:
[attr for attr in dir(field) if not attr.startswith('__')]
['check_bcs_present',
'check_connectivity',
'elem',
'elmap',
'endian',
'get_points',
'istep',
'lims',
'lr1',
'merge',
'nbc',
'ncurv',
'ndim',
'nel',
'offset_connectivity',
'time',
'update_ncurv',
'var',
'wdsz']
Here check_connectivity and merge are methods, and elem and elmap are data objects. The rest of the attributes store metadata. See pymech.core to understand what they imply.
print(field.endian, field.istep, field.lr1, field.nbc, field.ncurv, field.ndim, field.nel, field.time, field.var, field.wdsz)
little 10 (8, 8, 8) 0 [] 3 512 0.2 (3, 3, 1, 0, 0) 4
The elem attribute contains data of physical fields. It is an array of lists, with each array representing an element.
print("There are", field.nel, "elements in this file")
There are 512 elements in this file
pymech.core.Elem#
The raw arrays are stored as a list of pymech.core.Elem instances as pymech.core.HexaData’s elem attribute.
first_element = field.elem[0]
print("Type =", type(first_element))
print(first_element)
Type = <class 'pymech.core.Elem'>
<elem centered at [ 0.39269908 -0.98 0.19634954]>
Let us look at the attributes of an element
[attr for attr in dir(first_element) if not attr.startswith('__')]
['bcs',
'ccurv',
'centroid',
'curv',
'face_center',
'pos',
'pres',
'scal',
'smallest_edge',
'temp',
'vel']
Except for the following attributes of an element object
print(first_element.bcs, first_element.ccurv, first_element.curv)
[] ['', '', '', '', '', '', '', '', '', '', '', ''] [[0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0.]]
it contains large arrays
print("Shape of element velocity and pressure arrays = ", first_element.vel.shape, first_element.pres.shape)
Shape of element velocity and pressure arrays = (3, 8, 8, 8) (1, 8, 8, 8)
pymech.dataset#
from pymech.dataset import open_dataset
ds = open_dataset('channel3D_0.f00001')
This function loads the field file in a more convenient xarray dataset.
ds
<xarray.Dataset> Size: 15MB
Dimensions: (x: 64, y: 64, z: 64)
Coordinates:
* x (x) float64 512B 0.0 0.05037 0.1603 0.3105 ... 6.123 6.233 6.283
* y (y) float64 512B -1.0 -0.9974 -0.9918 ... 0.9918 0.9974 1.0
* z (z) float64 512B 0.0 0.02518 0.08017 0.1553 ... 3.061 3.116 3.142
time float64 8B 0.2
Data variables:
xmesh (z, y, x) float64 2MB 0.0 0.05037 0.1603 ... 6.123 6.233 6.283
ymesh (z, y, x) float64 2MB -1.0 -1.0 -1.0 -1.0 -1.0 ... 1.0 1.0 1.0 1.0
zmesh (z, y, x) float64 2MB 0.0 0.0 0.0 0.0 ... 3.142 3.142 3.142 3.142
ux (z, y, x) float64 2MB 0.0 0.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0 0.0
uy (z, y, x) float64 2MB 0.0 0.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0 0.0
uz (z, y, x) float64 2MB 0.0 0.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0 0.0
pressure (z, y, x) float64 2MB 0.004913 0.0467 0.0195 ... 0.03768 0.0761The dataset is more descriptive and useful for exploratory work, such as post-processing and plotting.
Computing statistics#
Calculate median for all variables
ds.median()
<xarray.Dataset> Size: 64B
Dimensions: ()
Coordinates:
time float64 8B 0.2
Data variables:
xmesh float64 8B 3.142
ymesh float64 8B 0.0
zmesh float64 8B 1.571
ux float64 8B 0.8375
uy float64 8B 0.0
uz float64 8B 0.0
pressure float64 8B 0.0001238Slicing#
Slice by index:
ds.ux.isel(z=32)
<xarray.DataArray 'ux' (y: 64, x: 64)> Size: 33kB
array([[0. , 0. , 0. , ..., 0. , 0. ,
0. ],
[0.006773 , 0.01130644, 0.01251847, ..., 0.01209086, 0.01235788,
0.006773 ],
[0.02467888, 0.03595118, 0.03948131, ..., 0.03794473, 0.03727177,
0.02467888],
...,
[0.03804531, 0.0281054 , 0.03863081, ..., 0.03893135, 0.04252424,
0.03804531],
[0.01240053, 0.00786791, 0.01261182, ..., 0.01296887, 0.01406241,
0.01240053],
[0. , 0. , 0. , ..., 0. , 0. ,
0. ]])
Coordinates:
* x (x) float64 512B 0.0 0.05037 0.1603 0.3105 ... 6.123 6.233 6.283
* y (y) float64 512B -1.0 -0.9974 -0.9918 -0.9842 ... 0.9918 0.9974 1.0
z float64 8B 1.571
time float64 8B 0.2It is also possible to slice by value using sel method
Visualizing#
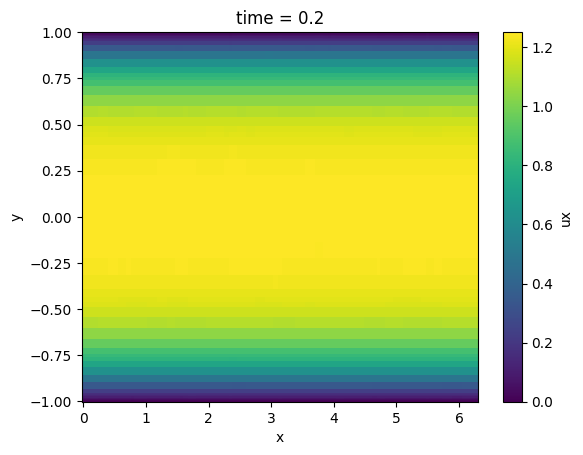
Average in spanwise (z) direction and plot velocity profile
ds.ux.mean('z').plot()
<matplotlib.collections.QuadMesh at 0x7f16516620d0>
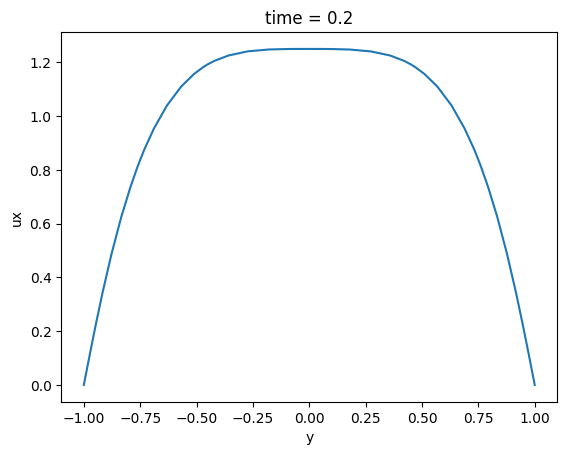
Average in both horizontal direction and plot 1D profile
ds_1d = ds.mean(['x', 'z'])
ds_1d.ux.plot()
[<matplotlib.lines.Line2D at 0x7f1651366520>]
It is also worth knowing that it is possible to:
Parallelize these operations using
ds.chunkmethod followed byds.computeOpen a multiple files into a single dataset using
pymech.dataset.open_mfdataset, optionally in parallel.
Read the xarray documentation to see how to use them.
pymech.meshtools#
This class helps working with Nek5000 meshes. It can modify existing meshes and generate new ones.
2D Mesh generation#
Boxes#
You can generate a rectangular box mesh like this:
import pymech.meshtools as mt
# box of size 5×1
xmin = 0
xmax = 5
ymin = 0
ymax = 1
# 20 × 10 elements resolution
nx = 20
ny = 10
# velocity inflow on the left
bc_inflow = ['v']
# outflow on the right
bc_outflow = ['O']
# wall at the bottom
bc_wall = ['W']
# free-stream (fixed vx and d(vy)/dy = 0) at the top
bc_freestream = ['ON']
box = mt.gen_box(
nx,
ny,
xmin,
xmax,
ymin,
ymax,
bcs_xmin=bc_inflow,
bcs_xmax=bc_outflow,
bcs_ymin=bc_wall,
bcs_ymax=bc_freestream,
)
It is also possible to start from a square box and map it to a different shape, or simply move the elements in the box to change the resolution locally. For example, this refines the resolution close to the wall such that the first element has a height of 0.025 instead of 0.1.
l0 = 0.025
alpha = mt.exponential_refinement_parameter(l0, ymax, ny)
# maps (x, y) -> (x', y')
def refinement_function(x, y):
iy = ny * y / ymax
return (x, l0 * (1 - alpha**iy)/(1 - alpha))
# This mapping doesn't introduce any curvature, so we don't need to encode any new curvature in the mesh
box = mt.map2D(box, refinement_function, curvature=False, boundary_curvature=False)
Other geometries#
Pymech also lets you generate a disc mesh. The following example generates a disc domain with a temperature field
# the disc will have radius 1
radius = 1
# centre box of 5×5 elements
n_centre = 5
# four side boxes of 6×5 elements
n_bl = 6
# the diagonal of the centre box will be half of the disc diametre
s_param = 0.5
# (x, y), (vx, vy), p, t, no passive scalars
variables = [2, 2, 1, 1, 0]
# boundary conditions: adiabatic wall
bcs_sides = ['W', 'I']
# generate the mesh
disc_mesh = mt.gen_circle(radius, s_param, n_centre, n_bl, var=variables, bc=bcs_sides)
Extrusion#
Pymech can extrude 2D meshes into 3D ones.
We can for example extrude the disc into a cylinder with velocity and temperature inlet at the bottom and outflow at the top:
# extrude in ten elements vertically between -1 and +1
z = [-1, -0.9, -0.7, -0.5, -0.25, 0, 0.25, 0.5, 0.7, 0.9, 1]
bc_inflow = ['v', 't']
bc_outflow = ['O', 'I']
# bc1 denotes the boundary conditions at z = -1, and bc2 at z = +1
cylinder_mesh = mt.extrude(disc_mesh, z, bc1=bc_inflow, bc2=bc_outflow)